Fudan's scientific research team has cooperated with scientists from many countries to build the most comprehensive global microbial gene catalog to date. In the early morning of December 16, Beijing time, the relevant research results were published in the main issue of Nature.
Microorganisms are ubiquitous in the earth, hidden in people's skin, intestines and soil, rivers, oceans and other environments, forming a complex microbiome community. They interact symbiotically with different hosts in different environments and become important factors affecting human health, disease development and ecological changes on the earth. Traditional microbiome research is based on different habitats such as human microorganisms and marine microorganisms, and it is impossible to describe the interconnection of microbial communities in different habitats from a global perspective.
Luis Pedro Coelho, a young researcher at the Institute of Brain-Like Research of Fudan University, Professor Zhao Xingming, and Professor Emeritus Peer Bork collaborated with scientists from Germany, Spain, the United States, the United Kingdom and other countries to study, based on the concept of the global microbiome, the microorganisms of different habitats on the earth as a unified system. Using artificial intelligence technology to mine 13,000 publicly available metagenomic samples, the most comprehensive Global Microbial Gene Catalog (GMGC) to date was constructed, an important step forward in global microbiome research. The study also found that most genes are habitat-specific, with genes that span multiple habitats predominantly enriched in antibiotic resistance genes and mobile genetic elements.
In the early morning of December 16, Beijing time, the relevant research result "Towards the biogeography of prokaryotic genes" was published in the main issue of Nature in the form of a long article, and Coelho was the first author and co-corresponding author of the paper.
The research has been funded by the EU's "Horizon 2020" Innovation Program, the National Key Research and Development Program, the National Natural Science Foundation of China, and the Shanghai Municipal Science and Technology Major Project of "Brain and Brain-like Intelligence Basic Transformation and Application Research".
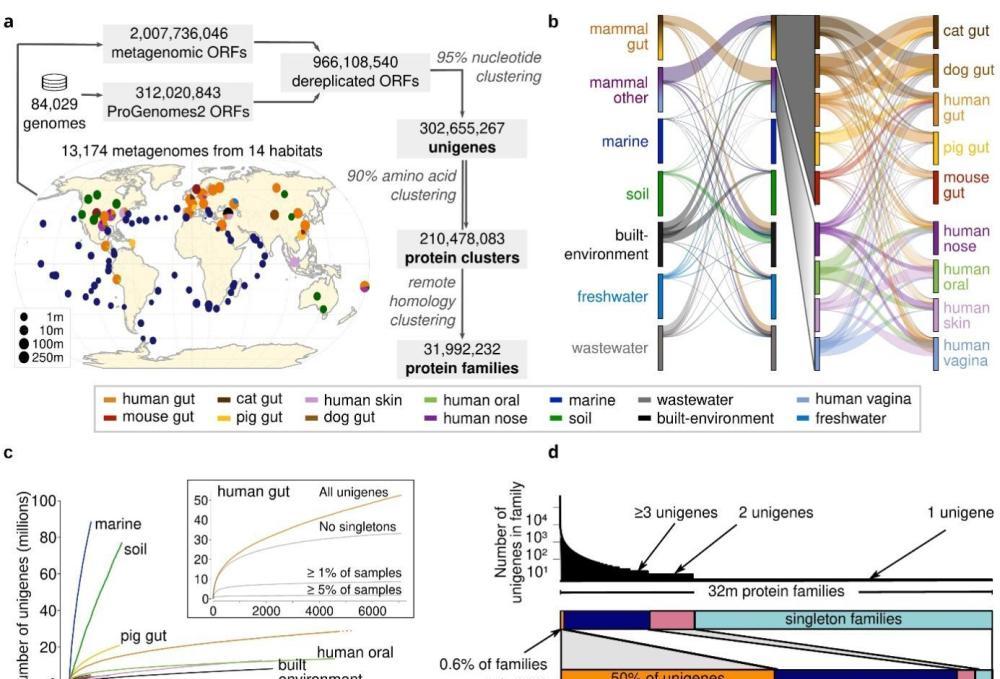
A global microbial gene catalog of 300 million prokaryote genes. The pictures in this article are provided by Fudan University
Uncover important associations between microbial genes and habitats
Gene catalogs are important for describing the species composition and functional characteristics of microbial communities. Since the European Molecular Biology Laboratory (EMBL) and BGI genetics constructed the first human gut microbial gene catalog in 2010, the emerging microbial gene catalog has provided important clues for the study of human physiology and disease.
The Global Microbial Gene Catalog (GMGC) covers the main habitats of 14 microorganisms, including the gut, mouth, skin, ocean, soil, etc., collects 13,174 publicly available high-quality metagenomics and 84,029 high-quality genomes, obtains genes containing 303 million species levels (95% nucleic acid consistent clustering), and constructs the most comprehensive global microbial gene catalog to date. It will provide important contributions to the study of earth ecology and human health.
Species-level unigenes may represent genes from multiple habitats . Multi-habitat genes may come from species that live in multiple habitats, or from mobile genetic elements that are transferred horizontally between genomes or across habitat boundaries. The study found that most genes are habitat-specific, which is consistent with the characteristics of microorganisms that tend to adapt to the environment; only 5.8% of the single gene clusters at the species level are multi-habitat genes, and multi-habitat genes are mainly enriched in antibiotic resistance genes and mobile genetic originals.
The researchers further studied the frequency of single gene clusters in the metagenomic and found that most single gene clusters are rare genes that appear less than 0.1% more frequently. The frequencies of single gene clusters follow a power law distribution under the neutral (or near-neutral) evolutionary hypothesis. In fact, while a lot of variation has been observed, most variation is not an adaptation to the environment, but is driven by so-called "neutral evolution": variation is only a random result, not a Darwinian choice.
These findings are important for understanding the emergence of antibiotic resistance, as well as the future development of antimicrobial drugs.
Seminar at the Institute of Brain-like Research of Fudan University
An international research team with a multidisciplinary blend
The biomedical artificial intelligence team of the Institute of Brain-like Intelligent Science and Technology of Fudan University focuses on the intersection of artificial intelligence and biomedicine, and has introduced a group of outstanding scholars at home and abroad since 2018. The team has different discipline backgrounds such as computer, mathematics, biology, physics, etc., and has a strong international atmosphere. In recent years, around the characteristics of biomedical big data, a series of artificial intelligence algorithms have been developed, which have been successfully applied to scenarios such as brain-intestinal axis, brain development and brain diseases, and the relevant work has been published in journals such as Nature, Science, cell and so on.
The paper's first author, Coelho, joined Fudan full-time in 2018, having previously focused on the analysis of microbial communities using metagenomic and microscopic imaging techniques. He cooperated with multiple research groups such as the biological big data team and the computational biology team of the Institute, participated in the construction of the Key Laboratory of Computational Neuroscience and Brain-like Intelligence of the Ministry of Education, participated in a number of major research projects such as biomedical big data, brain science and brain-like research as the backbone of the project, and focused on the use of big data methods to engage in microbiome and precision medicine and other related research.
As the head of the biomedical artificial intelligence team of the Brain-like Research Institute, Professor Zhao Xingming introduced that the study constructs a catalog of microbial genes from a global perspective, which plays an important role in understanding the relationship between microorganisms and human health. In the next step, the team will further cooperate with domestic and foreign research institutes and clinical medical institutions based on the developed gene catalog to explore the influence of microorganisms, including human gut microbes, human life health, brain cognition and behavior.