About Dr.X:
Dr. X is a professional knowledge sharing column initiated by the Jintai Technology Doctoral Group, which aims to transmit and share the cutting-edge technology and R&D dynamics of global AI drug research and development to the biomedical industry, promote the awareness of AI drug research and development among drug development practitioners, and promote the application of AI and other cutting-edge technologies in the field of drug research and development.
The concept of Synthetic Lethality was first proposed by American genetic scientist Calvin Bridge in 1922 and officially named by Theodore Dobzhansky in 1946. A simple understanding of synthetic death is that two genes in a cell, when either one alone mutates or does not function, the cell can survive; but when both are functionally defunctional at the same time, it will lead to cell death. In recent years, synthetic death has played an increasingly important role in the precision treatment of tumors.
Based on the special mechanism of synthetic death, this paper will discuss the application and value of synthetic death in the field of tumors through several small questions.
Why choose synthetic lethal therapy?
Targeted drugs are known to specifically bind to and act on key functional proteins in tumor cells, causing their death, but to date, there have been relatively few tumor targets that can be drugged. Using synthetic lethal mechanisms, it is possible to indirectly target tumor-specific mutations and conduct research and development on those "incurable" tumor mutation genes. At the same time, synthetic lethal therapy also shows some potential to overcome the drug resistance problem caused by new mutations in targeted therapy.
The main application scenario of synthetic death?
Targeted DNA damage repair (DDR) is currently the main practice of "synthetic death". A variety of endogenous and exogenous factors can continuously induce DNA damage, such as base damage, single-strand breaks (SSBs), double-strand breaks (DSBs), etc., of which double-strand breaks have the most lethal effect on cells. For DNA damage, cells have evolved a series of DNA damage repair pathways to maintain gene stability, including homologous recombinant repair pathways (HRR), non-homologous end-linking pathways (NHEJ), base excision repair (BER), mismatch repair pathways (MMR), and so on.
There is interdependence between DDR pathways, and when a genetic mutation in DNA causes damage to some DDR components, the survival of cells will depend on other DDR mechanisms. At this time, if the paired gene of the mutated gene is found and other DDR pathways are inhibited, the DNA damage of the tumor cells will not be repaired, which will lead to death.
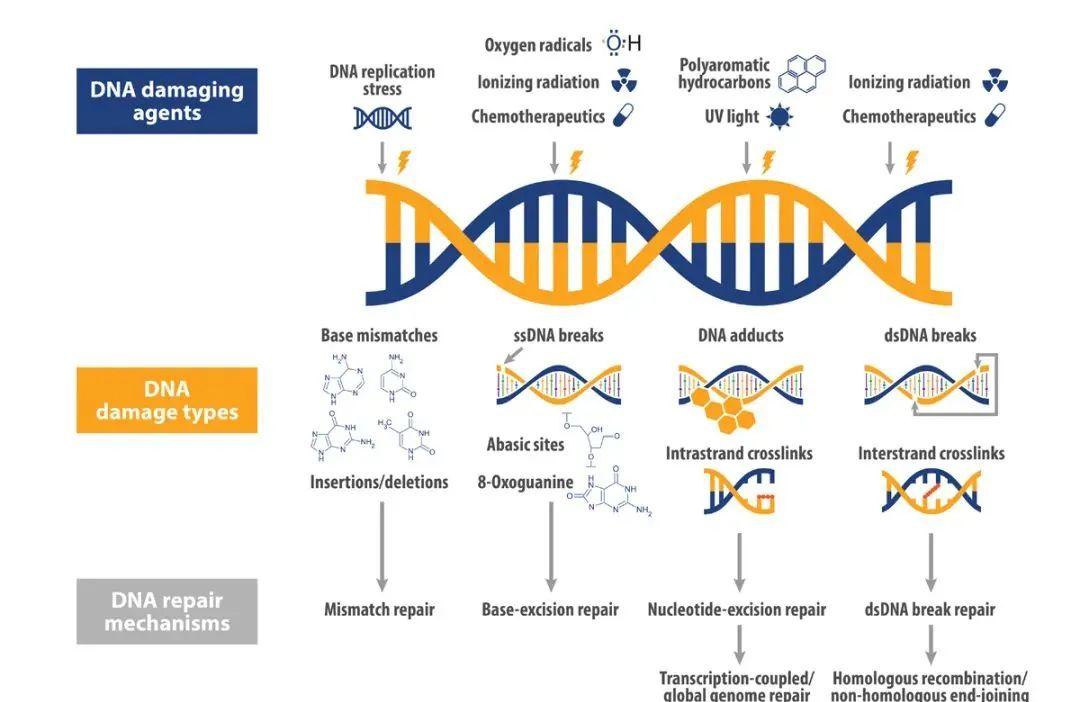
Figure 1 DNA Damage Repair Pathway [1]
What are the pairs of lethal genes synthesized in research?
Marketed synthetic lethal inhibitor: PARPi
The most successful and only market-proven synthetic lethal gene pair is BRCA/PARP. Tumors with mutations in the BRCA1/2 gene often have homologous recombinant defects (HRDs), resulting in defects in the cellular DNA double-strand break damage repair (DSB) pathway; additional blocking of a PARP-regulated DDR pathway, base excision repair (BER), will cause DNA damage to accumulate and reach critical numbers, killing cells [2].
Figure 2 Mechanism of PARP resistance [3]
PARP inhibitors have shown good efficacy in ovarian, breast, prostate cancer and other cancers. For example, at a recent ASCO-GU meeting, the results of the PROpel study were reported orally: Olapally plus abitulone in a placebo combination group resulted in a significant rPFS benefit of 8.2 months (24.8 months vs 16.6 months; HR 0.66, 95% CI 0.54-0.81; P< 0.0001) for patients treated in the first line of mCRPC, regardless of HRR status.
Figure 3 PROpel Primary Study Endpoint rPFS, 2022 ASCO-GU
In the post-PRAPi era, what other potential synthetic lethal targets are worth looking forward to
The remarkable efficacy of PARPi has allowed everyone to see the wide application prospect of synthetic death in the field of tumors, but due to BRCA1/2 function recovery, non-BRCA1-dependent HR recovery, and recovery of replication fork stability[ 3 ] , drug resistance problems have also emerged. Inhibiting DDR compensatory pathways, finding new synthetic lethal interactions, and precise targets are also points for scientists to further explore and think.
At present, synthetic lethal research outside PARPi is still in the exploratory stage, mainly preclinical and Phase I./II clinical trials. By analyzing the targets of the current research, it can be found that there are two main aspects of the research on synthetic lethality:
The first is to look for other pairable synthetic lethal targets, hoping to find the next "PARPi", which is also the main focus of everyone.
Here are some of the genes that have been identified that may have a "synthetic-to-lethal" relationship:
ATR and ATM:
ATR and ATM are signaling kinases that sense DNA damage and are the main activators of DDR. After ATR function is lost, cancer cells will rely on the ATR pathway; inhibition of ATR can achieve a killing effect on cancer cells, which indicates a synthetic lethal relationship between ATR and ATM; in addition, ATR inhibition may be compensated by DNA-PK function.
WEE1 vs TP53:
WEE1 is part of the serine/threonine protein kinase family that inhibits the activity of CDK kinases, thereby inhibiting cells from entering mitosis; Wee1 and P53 form a pair of synthetic lethal targets to some extent. In 2021, data disclosed at the AACR Annual Meeting validated that tumors with TP53 mutations have some sensitivity to WEE1 inhibitors.
RAD51 and AID:
RAD51 is a highly fidelity recombinant enzyme involved in maintaining gene stability, DNA damage response, and cell cycle regulation, playing a central role in homologous recombination (HR) processes. A 2021 ASCO study (Abstract #3006) showed that the RAD51 inhibitor CYT-0851 selectively induced aid-highly expressing tumor cells to damage DNA accumulation.
PRMT5 vs MTAP:
As a classic epigenetic target, PRMT5 also shows some potential in "synthetic lethality". PRMT5 knockout selectively kills MTAP to remove tumor cells without affecting normal cells.
In addition, there are chK1/2, DNA-PK, POLθ, USP1 and other targets are also being studied, which are not introduced for reasons of length.
The second is to try to solve the problem of PARPi drug resistance by inhibiting other compensatory DDR pathways.
This aspect was reported in both ASCO and AACR 2021. At ASCO 2021, Wee1 inhibitors (Abstract #5505) and ATR inhibitors (Abstract #5516) both showed potential to overcome RESISTANCE to PARP1/2 inhibitors; the official AACR blog also mentioned that POLθ inhibitors and ATR inhibitors may help overcome resistance to PARP inhibitors [2].
Figure 4 Synthetic lethal targets based on DNA damage repair and drugs under development [4]
How to discover synthetic lethal gene pairings?
PARP inhibitors have illustrated the broad promise of synthetic death in the field of tumor treatment, and other synthetic death research is in full swing, but the DDR signaling pathway is very complex, and how to find the appropriate synthetic death-related gene mutation is the difficulty.
With the development and progress of gene sequencing technology, CRISPR-based phenotypic screening technology has the advantages of high throughput, small bias and high accuracy in discovering new drug action targets. CRISPR accelerates the development of DDR inhibitors other than PARP by knocking out specific genes from tumor cells, observing the survival status of tumor cells, and finding potential lethal genes. At the same time, CRISPR phenotype screening combined with machine learning algorithms can reduce dimensions and cluster data in complex phenotypes, especially unsupervised machine learning, to discover new unknown targets and mechanisms of action, and gain more and more consensus [6]; scholars at the University of Cambridge have developed a machine learning algorithm based on systematic CRISPR screening to accurately screen tumors with mismatch repair defects that are more sensitive to immunotherapy [7].
Fig. 5 CRISPR screening experimental design[5]
Algorithm optimization is particularly important in expanding new mechanisms and new target discoveries based on phenotypic screening. The complexity of the biological organism determines that the interpretation of biological data cannot be completely replaced by algorithms, so the development of more humane and easy-to-use algorithms, and the focus on human-machine (algorithm) collaboration, high-throughput data analysis with high experience but without losing objectives will be the focus of the next step of optimization.
Showcase
Through the above few small questions, we roughly discussed the application and value of synthetic death, and also saw the broad space of synthetic death in the field of tumor treatment. Compared with traditional chemotherapy and radiotherapy, synthetic lethality is still very "young", and there are many problems that need to be jointly explored and solved by all sectors of society, such as how to further clarify the interdependence between DDR signaling pathways, what new synthetic lethal possibilities are outside of DDR, and how to more effectively solve PARPi resistance. It is believed that in the future, with the development of technology and the continuous research of researchers, as well as the empowerment of cutting-edge technologies such as machine learning, synthetic lethal therapy will usher in a breakthrough to provide patients with more effective and accurate treatment solutions.
bibliography:
[1] https://blog.crownbio.com/dna-damage-response#_
[2]https://www.aacr.org/blog/2021/12/06/molecular-targets-2021-examining-synthetic-lethality-beyond-parp-inhibitors/
[3] Dias MP, et al. Nat Rev Clin Oncol. 2021; doi:10.1038/s41571-021-00532-x
[4] P. Pilié, Nat Rev Clin Oncol. 2019
[5] Bock, et al. Nat Rev Methods Primers 2, 8 (2022). https://doi.org/10.1038/s43586-021-00093-4
[6]https://www.drugtargetreview.com/article/44422/phenotypic-profiling-in-drug-discovery/
[7] Zou, X.,et al. Nat Cancer 2, 643–657 (2021). https://doi.org/10.1038/s43018-021-00200-0
This article is for academic exchange only, the source network, invasion and deletion
About Jintai Technology
Jintai Technology is a drug research and development company empowered by quantum physics and artificial intelligence, which is committed to achieving industry innovation in drug research and development by improving the speed, scale, innovation and success rate of drug research and development. As a company based in China and the United States and serving the world, Jintai Technology always insists on exploring the best solutions to make full use of cutting-edge R&D and computing resources to maximize the needs of customers and partners.
Jintai Technology's smart drug R&D platform integrates digital R&D tools based on cloud supercomputing with advanced experimental capabilities to form a R&D system in which high-precision prediction and targeted experiments corroborate and guide each other. As one of the world's pioneering artificial intelligence drug R&D companies, Jintai Technology has established a complete set of R&D iteration processes closely integrated with quantum physics dry laboratories and advanced wet laboratories, challenging the efficiency bottleneck of traditional R&D and enabling new drug research and development to achieve breakthroughs in innovation speed and scale.
For business/event inquiries, please contact: [email protected]