When it comes to machine learning, it is necessary to mention programming, configuration environment, and model training, which makes friends who have needs but no programming foundation prohibitive.
Weka, developed based on JAVA, is a good open source software for machine learning and data mining. In the previous article (high-quality cutouts of Arabidopsis in top journals, no PS use this, just click the mouse) introduced an artifact ImageJ, and Weka is also integrated as a plug-in in the software.
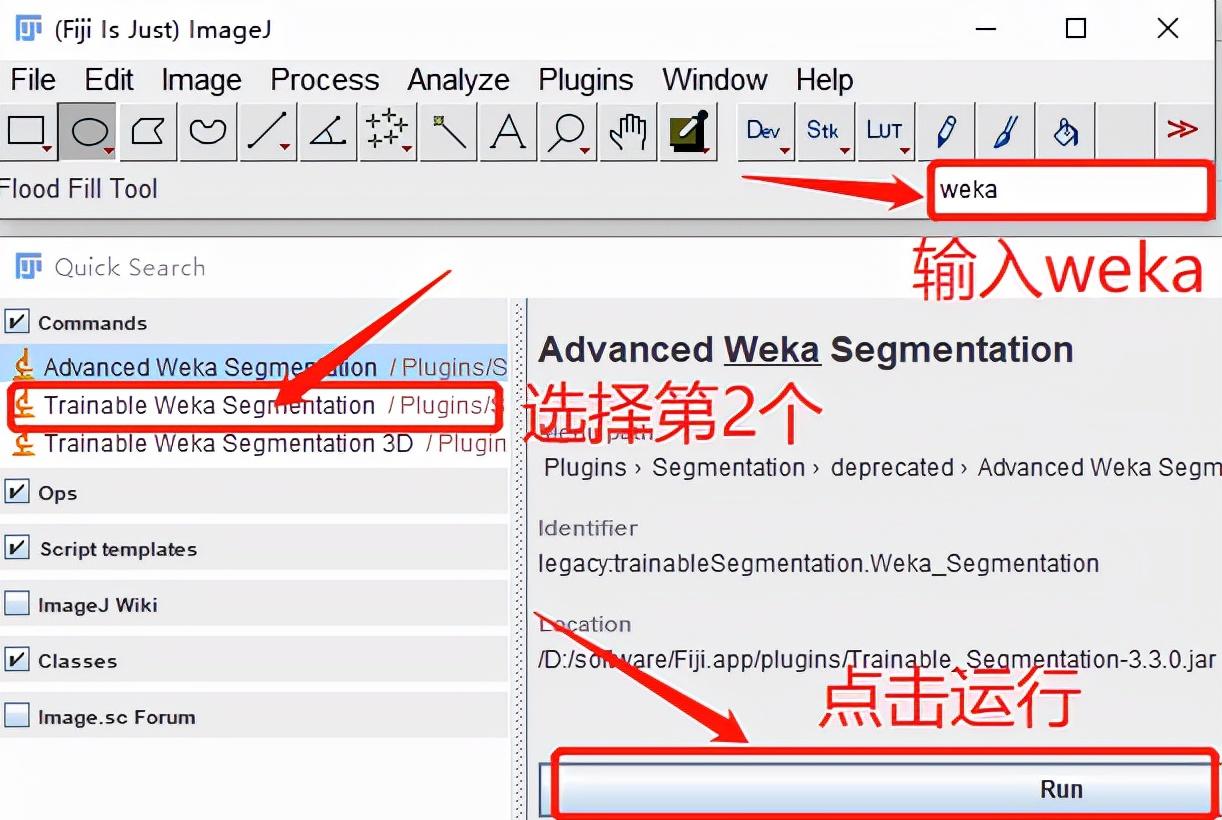
Immediately after the software pop-up prompt to open the picture, we trained this time to identify the spore model, as shown in the following figure:
Once you have selected the image, you will be able to enter the WEKA interface.
The interface includes:
Training bar: Train classifier
Options bar: Load classifier, Load Data, Create new class, Settings
Tab bar: Add different categories (here the classification defaults to two, which can be set in detail in Settings)
We enter the settings where we can rename the categories, which prevents confusion.
<h1 class="pgc-h-arrow-right" data-track="80" > model training</h1>
Next we start training the model
First select the Shape Selection tool on the ImageJ panel, circle the spores and fill in the spore classification (red module), the same classification background (green module). After selecting all, click train classifier to wait for the machine to finish training.
After a short wait, we found that all the spores were marked red with a green background.
Model training is not bad, but it still needs to be fine-tuned. If the adjacent spores are circled together, we need to separate them, so this time we use dashes to divide and add to the background label, and then train again.
After training the model again, the model at this time is better than the last time, and if you are not satisfied, you can continue to fine-tune.
<h1 class="pgc-h-arrow-right" data-track="86" > count</h1>
Select Create result after training
The result after clicking is as follows:
At this time, only these few steps are required:
1、Image→Type→8 bit
2、Image→Adjust→Threshold
3. Drag the pulley to adjust to the white background Black spores as shown below
4、Process→Filters→Median
5、Process→Binary→Watershed
6. Analyze→Analyze Particles (Note: Because there are spores on the boundary, do not check Exclude on edges)
The counting results are as follows:
<h1 class="pgc-h-arrow-right" data-track="97" > save and call the model</h1>
To save the model:
We go back to the WEKA interface again and click on Save classifier, the file format is classifier.model
Call model
Use WEKA to open a new spore image and load the model directly (Load classifier), then directly create result to analyze the picture, and then repeat the number of statistics operation process.
At last
Peng Duanshu of the Qing Dynasty said in "A Show of Sons and Nephews for Learning", "Is it difficult or easy to do things in the world? For this, it is easy to be difficult; if it is not, it is also difficult to be easy. Is it difficult to learn human beings? If you learn, it is easy to learn, and if you don't learn, it is easy to learn. ”
Good luck to everyone, SCI sent to the soft hand ~