論文
GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC Department of Botany and Plant Sciences, University of California, Riverside Bioinformatics
GDC: The Genomic Data Commons
基本功能
- 資料下載下傳
- ceRNA網絡分析
- 差異表達分析
- 功能富集分析
- 生存分析
- 資料可視化 火山圖、熱圖、GO富集分析結果、KEGG富集分析結果等
接下來重複幫助文檔中的例子
幫助文檔連結 http://bioconductor.org/packages/devel/bioc/vignettes/GDCRNATools/inst/doc/GDCRNATools.html
library(GDCRNATools)
project<-'TCGA-CHOL'
rnadir<-paste(project,'RNAseq',sep='/')
mirdir<-paste(project,'miRNAs',sep="/")
gdcRNADownload(project.id = 'TCGA-CHOL',
data.type = 'RNAseq',
write.manifest = F,
method = 'gdc-client',
directory = rnadir)
複制
在linux系統中重複到這一步的時候遇到報錯 ImportError: /lib64/libc.so.6: version `GLIBC_2.18' not found (required by /tmp/_MEIylVP0W/libstdc++
我的解決辦法是把它預設下載下傳的gdc-client_v1.3.0替換掉,我換成gdc-client_v1.5.0,下載下傳位址是https://gdc.cancer.gov/access-data/gdc-data-transfer-tool
gdcRNADownload(project.id = 'TCGA-CHOL',
data.type = 'miRNAs',
write.manifest = F,
method = 'gdc-client',
directory = mirdir)
clinicaldir<-paste(project,'Clinical',sep='/')
gdcClinicalDownload(project.id = 'TCGA-CHOL',
write.manifest = F,
method='gdc-client',
directory = clinicaldir)
metaMatrix.RNA<-gdcParseMetadata(project.id = 'TCGA-CHOL',
data.type = 'RNAseq',
write.meta = F)
metaMatrix.RNA<-gdcFilterDuplicate(metaMatrix.RNA)
metaMatrix.RNA<-gdcFilterSampleType(metaMatrix.RNA)
metaMatrix.MIR<-gdcParseMetadata(project.id = 'TCGA-CHOL',
data.type = 'miRNAs',
write.meta = F)
metaMatrix.MIR
metaMatrix.MIR<-gdcFilterDuplicate(metaMatrix.MIR)
metaMatrix.MIR<-gdcFilterSampleType(metaMatrix.MIR)
複制
擷取表達矩陣
rnaCounts<-gdcRNAMerge(metadata = metaMatrix.RNA,
path = rnadir,
organized = FALSE,
data.type = 'RNAseq')
mirCounts<-gdcRNAMerge(metadata = metaMatrix.MIR,
path = mirdir,
organized = FALSE,
rnaCounts[1:5,1:5]
mirCounts[1:5,1:5]
複制
标準化表達資料
rnaExpr<-gdcVoomNormalization(counts=rnaCounts,filter=F)
mirExpr<-gdcVoomNormalization(counts=mirCounts,filter=F)
rnaExpr[1:5,1:5]
mirExpr[1:5,1:5]
複制
差異表達分析
DEGAll<-gdcDEAnalysis(counts = rnaCounts,
group=metaMatrix.RNA$sample_type,
comparison = 'PrimaryTumor-SolidTissueNormal',
method='limma')
deALL<-gdcDEReport(deg=DEGAll,gene.type = 'all')
deLNC<-gdcDEReport(deg=DEGAll,gene.type='long_non_coding')
dePC<-gdcDEReport(deg=DEGAll,gene.type = 'protein_coding')
複制
記下來是資料可視化展示
柱形圖展示差異表達的基因類型
gdcBarPlot(deg=deALL,angle = 45,data.type = 'RNAseq')
複制
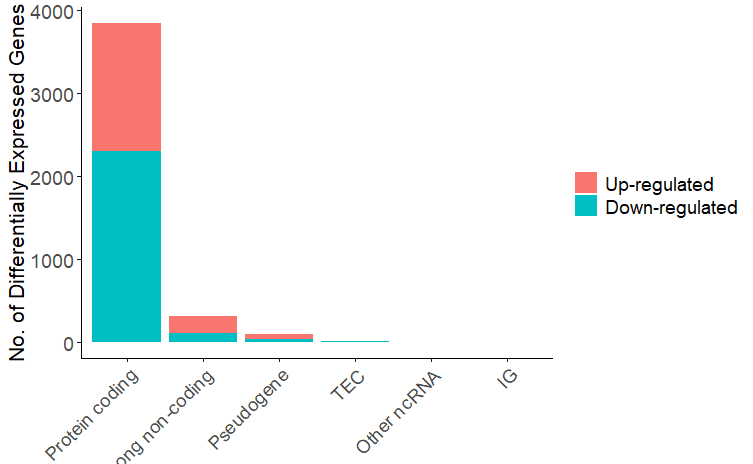
image.png
這裡TEC和IG分别是啥?
長鍊非編碼RNA的差異表達火山圖
gdcVolcanoPlot(deLNC)
複制
熱圖
degName<-rownames(deLNC)
gdcHeatmap(deg.id = degName,metadata = metaMatrix.RNA,rna.expr = rnaExpr)
複制
image.png
富集分析
enrichOutput<-gdcEnrichAnalysis(gene=rownames(deALL),
simplify=T)
gdcEnrichPlot(enrichOutput,type='bar',category = 'GO',num.terms = 10)
複制
畫圖的時候遇到報錯 Error in .Call.graphics(C_palette2, .Call(C_palette2, NULL)) : invalid graphics state 不知道原因出在哪裡,但是儲存到本地沒問題
pdf(file="../goenrich.pdf",width = 15,height = 15)
gdcEnrichPlot(enrichOutput,type='bar',category = 'GO',num.terms = 10)
dev.off()
複制
image.png
ceRNA網絡
ceOUtput<-gdcCEAnalysis(lnc=rownames(deLNC),
pc=rownames(dePC),
lnc.targets = 'starBase',
pc.targets = 'starBase',
rna.expr = rnaExpr,
mir.expr = mirExpr)
edges<-gdcExportNetwork(ceNetwork = ceOutput2,net='edges')
nodes<-gdcExportNetwork(ceNetwork = ceOutput2,net='nodes')
write.table(edges,file='edges.txt',sep='\t',quote=F)
write.table(nodes,file="nodes.txt",sep="\t",quote=F)
複制
最後生成了兩個檔案,如何用cytoscape可視化這兩個檔案我暫時還不知道如何實作。
今天就先到這裡了。